Introduction
Streptococcus iniae is a gram-positive and significant bacterial pathogen of farmed fish worldwide including tilapia (Oreochromis sp.). The negative impacts in fish production caused by S. iniae have been estimated at more than $100 million (Shoemaker, Klesius, and Evans 2001; Shoemaker, Xu, and Soto 2017), although the current economic impact is not known. These large losses are due to a global distribution and an ability of S. iniae to cause disease in a large range of fish hosts (Agnew and Barnes 2007).
Gross signs of streptococcosis caused by S. iniae may include lethargy, dark coloration, spiral swimming behavior, unilateral or bilateral exophthalmia, corneal opacity, pustules of the caudal peduncle and jaw, and ascites fluid in the abdomen (Perera et al. 1994; Camus 2001; Chen, Chao, and Bowser 2007; Ortega et al. 2018). Streptococcus iniae infections are typically systemic and it is common to observe the bacterium free and/or attached to red blood cells in blood smears (Perera, Fiske, and Johnson 1998). The bacterium is commonly isolated from the brain and other internal organs of diseased fish (Ortega et al. 2018). Reported histological findings in infected fish include exudative meningoencephalitis, fibrinous to necrotizing splenitis, infiltration of mononuclear cells into several vital organs (heart, liver, kidney, spleen), and multiorgan hemorrhage (Camus 2001; Chen, Chao, and Bowser 2007). Intracellular and extracellular gram-positive cocci are often observed in the affected organs (Camus 2001; Chen, Chao, and Bowser 2007).
While treatment of streptococcosis is possible with judicious use of antibiotics (Gaunt et al. 2010; Gaikowski et al. 2014), prevention is preferred. Numerous preventative measures have been implemented and/or tested with various degrees of success including vaccination, probiotics, and targeted feed additives (Agnew and Barnes 2007; Maulu et al. 2021; Heckman et al. 2022). Another potential method that has gained interest is selective breeding for disease resistance. In 2014, our collaborative research team began assessing the feasibility to selectively breed Nile tilapia for disease resistance. Initial research focused on resistance to S. iniae (LaFrentz et al. 2016) and has since expanded to S. agalactiae (Shoemaker et al. 2017; LaFrentz et al. 2020), Francisella orientalis (Shoemaker et al. 2022), and tilapia lake virus (unpublished).
In the initial research with S. iniae, challenge of 143 families of Nile tilapia resulted in a high variation for mean survival of the families, ranging from 0 to 100%, and moderately high estimates of heritability were obtained demonstrating potential to improve this trait through selective breeding (LaFrentz et al. 2016). The ability to use family-based selection to improve disease resistance was confirmed via assortative mating that demonstrated tilapia selected for resistance had high survival (mean survival 88%) and those selected for susceptibility had low survival (mean survival 10%) following S. iniae challenge (Shoemaker et al. 2017). During these experiments, DNA samples were taken from fish for the purpose of a genome-wide association study (GWAS) to identify molecular markers (i.e., quantitative trait loci, QTL) linked to resistance to S. iniae. This research resulted in the identification of four markers significantly associated with resistance, explaining 12-26% of the genetic variance (Vela-Avitúa et al. 2023). The usefulness of the markers was confirmed by challenging offspring of brood tilapia classified as resistant or susceptible based on the markers in which offspring from resistant fish resulted in 1% mortality compared to 73% mortality for offspring from susceptible fish (Vela-Avitúa et al. 2023).
Although positive gains in resistance to S. iniae have been achieved, the underlying mechanisms involved are unknown. The availability of tilapia with robust disease resistance or susceptibility provides the opportunity to begin to address this knowledge gap. The objective of this work was to describe differences in mortality and pathology between strains of Nile tilapia resistant or susceptible to S. iniae.
Materials and Methods
Fish
Nile tilapia, O. niloticus, sires and dams from G7B1 (LaFrentz et al. 2020), that were previously genotyped as “resistant” or “susceptible” based on four quantitative trait loci (Vela-Avitúa et al. 2023), were used to produce the fish in this experiment. The two groups of tilapia were shipped to the Aquatic Animal Health Research Unit (AAHRU) as fry (~ 300 fish per group) and reared in two separate (400 L) tanks supplied with flow through dechlorinated municipal water at ~ 28 °C with supplemental aeration provided by air stones. Fish were fed appropriately sized Rangen (Buhl, ID, USA) fish food at a rate of ~14% body weight per day as fry and the feed rate was sequentially decreased to 3-4% once fish reached 50 g average weight. Stocking densities and flow rates were periodically adjusted to ensure proper water quality. Dissolved oxygen and temperature were checked daily and averaged 6.78 ± 1.03 ppm and 27.3 ± 0.77°C, respectively. Concentrations of ammonia, nitrite, and nitrate were checked twice a week and were always below 0.5 ppm, 0.5 ppm, and 20 ppm, respectively. For bacterial challenges, fish were transferred to 57 L tanks supplied with 0.5 L min-1 of the same water source indicated above. All procedures utilizing fish were approved by the USDA-ARS AAHRU Institutional Animal Care and Use Committee.
Bacteria and challenge
Two bacterial challenges were conducted, one to verify the resistant or susceptible phenotypes, and the second to harvest tissues pre- and post-challenge for histological analyses. For both challenges, an archived stock of S. iniae (ARS-98-60) generated from passage through Nile tilapia was used (LaFrentz et al. 2016; Shoemaker et al. 2017). Briefly, replicate samples of bacteria were grown overnight in tryptic soy broth (TSB) to optical densities at 540 nm of ~ 1.3 and then diluted 1:10 and 1:20 with TSB. The number of colony-forming units (cfu) mL−1 was determined by spread plating 100 μL volumes of 10-fold serial dilutions (in duplicate) onto sheep blood agar (SBA; Remel, Lenexa, KS, USA). Plates were incubated for 48 h at 28 °C, and colonies were counted and averaged to enumerate the cfu mL−1 using standard procedures.
To verify the phenotypes, tilapia (average weight, 48 g) were anesthetized with sodium bicarbonate buffered Tricaine-S (Syndel) and challenged by intraperitoneal injection of 100 µL suspension containing 1.48 × 107 cfu or 7.4 × 106 cfu. Two groups of 10 susceptible tilapia were injected with either the high or lower S. iniae dose, a single group of 10 resistant tilapia were injected with only the high dose, and a single group of 10 susceptible tilapia were mock challenged by injection of 100 µL sterile TSB. The second challenge conducted for histological sampling used tilapia with a mean weight of 100 g. A single group of 20 susceptible and a single group of 20 resistant tilapia were similarly injected with 100 µL volume corresponding to a S. iniae dose of 3.4 × 106 cfu fish-1. Feed was withheld from fish the day of the challenge, and following challenge, fish were placed into 57 L tanks supplied with 0.5 L min-1 flow through ~ 27.5 °C water and supplemental aeration was provided with air stones. Tanks were observed for behavior and morbidity/mortality at least twice daily. Dead fish and terminally moribund fish (euthanized with an overdose of Tricaine-S buffered 1:1 with sodium bicarbonate) were removed and recorded. Fish were examined for clinical signs of streptococcosis, and reisolation of S. iniae was attempted on ~ 20% of the daily mortalities from each tank by culturing brain tissue onto SBA. Reisolation was considered positive if at least one beta-hemolytic colony was visualized on plates after incubation for 48 h at 28°C.
Tissue sampling and histology
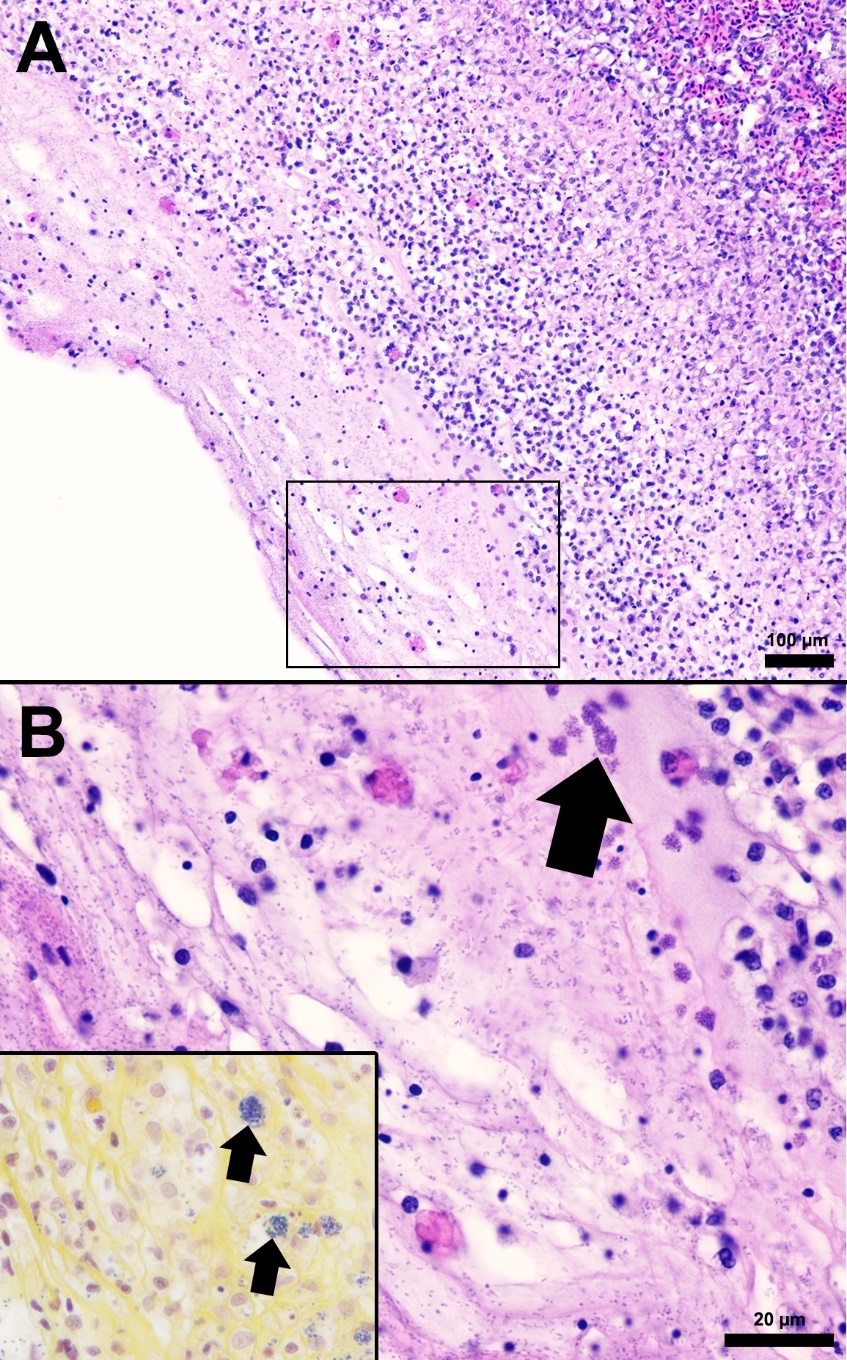
Resistant and susceptible tilapia were evaluated for microscopic pathology prior to S. iniae challenge and 24 h post-challenge. Five fish were sampled from each group at both timepoints. Fish were euthanized with sodium bicarbonate buffered Tricaine-S and necropsy was immediately performed to collect liver, spleen, anterior kidney, and whole brain tissues. Tissues were placed into ~40 mL of 10% neutral buffered formalin for 24 h and then transferred to ~40 mL of 70% isopropanol until processed for histological analyses. Tissues were placed into cassettes labeled with their treatment group; R = resistant; S = susceptible; the numbers refer to the sample collection timepoint 0 or 24 h post-challenge. The spleen and kidney segments were transferred in total, while the brain was bisected through the sagittal plane (if it was large enough, two brains from S0 tilapia remained whole) and a transverse section of liver was made. All tissues were routinely processed overnight, sectioned at 4 µm, and stained with a routine Gill II Hematoxylin and Eosin-Y protocol (Kiernan 2015; Jones 2020). McDonald’s Gram-stain kit (StatLab, Mckinney TX) was used on select sections to confirm Gram designation of bacteria. All sections were evaluated using a Nikon Eclipse 80i microscopic (Nikon, Tokyo Japan). Representative photomicrographs of normal and abnormal organ morphology were taken using an Amscope, MU1803-HS camera (Amscope, Irvine CA). All sections were evaluated by a board-certified veterinary pathologist blind to the experimental group designations. Lesions in each evaluated organ were subjectively delineated according to the severity of the pathological changes using a 4-tier system: minimal, mild, moderate, and severe.
Statistical analysis
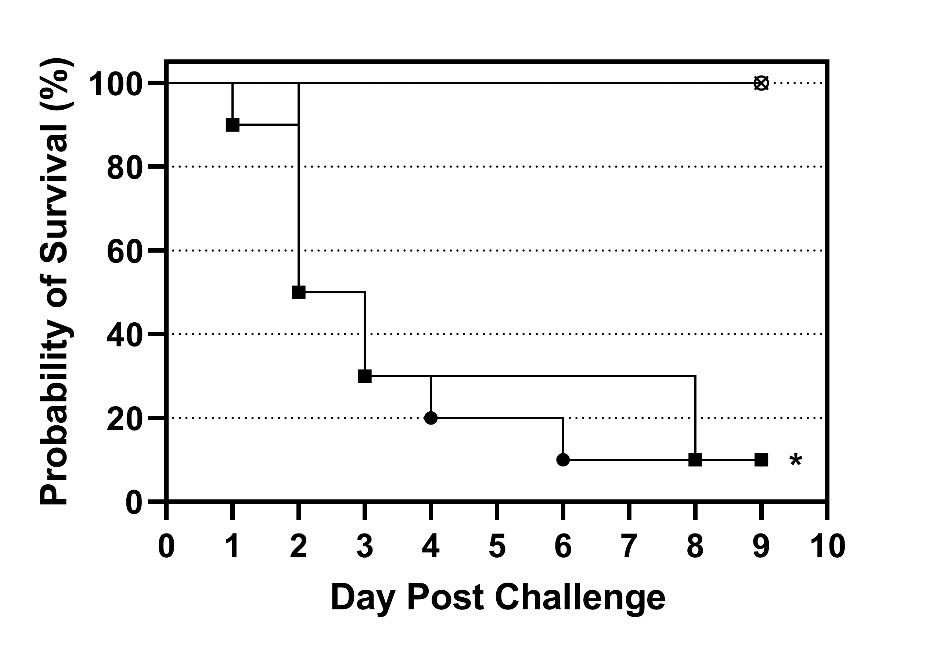
Survivor fractions for each group were calculated using the product limit (Kaplan–Meier) method with GraphPad Prism (version 9.3.1, GraphPad Software, LLC, San Diego, CA). Survival curves for the susceptible tilapia at both challenge doses were compared to the survival curve of the resistant tilapia at the high challenge dose using a log-rank test (Mantel-Cox). Thus, two pairwise comparisons were made, and differences were considered significant when P < 0.025 (Bonferroni correction; α = 0.05/2).
Results and Discussion
Through our selective breeding program, we have been able to successfully breed tilapia for resistance to S. iniae using MAS, and the resistant and susceptible phenotypes provides a unique opportunity to conduct research to discover differences in pathology and attempt to close the gap in the knowledge regarding the host mechanisms involved.
Experimental challenge of the resistant and susceptible tilapia confirmed our previous results demonstrating the resistant phenotype (Vela-Avitúa et al. 2023). The survival curves of susceptible tilapia challenged with both doses of S. iniae were similar with most of the mortality occurring between 1 and 3 d post challenge and final survival of both groups was 10% (Figure 1). In contrast, the resistant tilapia challenged with the high dose exhibited 100% survival and the survival curve was significantly (P < 0.025) different than the survival curves of the susceptible fish (Figure 1). No mortality occurred in the mock challenged group. Beta-hemolytic colonies characteristic of S. iniae were reisolated from 100% (8/8) of fish examined. There were also remarkable differences in behavior between the susceptible and resistant fish following challenge. The susceptible fish were lethargic and did not feed 24 h post challenge, whereas the resistant fish exhibited robust feeding behavior throughout the challenge. Due to the high mortality (50%) in the susceptible tilapia in less than 48 h, tilapia were sampled at 24 h post challenge for histologic analysis.
Following the confirmation of the phenotypes, resistant and susceptible fish were experimentally challenged with S. iniae and tissues (liver, spleen, anterior kidney, and brain) were collected for pathology. Examination of the pre-challenge susceptible and resistant tilapia (S0 and R0) samples did not identify any significant microscopic difference between the morphological characteristics of the liver, spleen, anterior kidney, and brain (Table 1). However, major differences were present in spleen, brain, anterior kidney, and liver tissues when comparing the S24 tilapia with all other groups (S0, R0, R24). Microscopic changes consistent with S. iniae infection were observed in the spleen, anterior kidney, and meninges of the S24 tilapia (Table 1). Similar to previous reports, the lesions in the affected fish were characterized by histiocytic to fibrinohistiocytic inflammation of the spleen, kidney, and meninges (Perera, Fiske, and Johnson 1998; Camus 2001; Chen, Chao, and Bowser 2007; Baums et al. 2013). In contrast, the resistant fish displayed few acute histopathological changes and no microscopically visible gram-positive cocci were observed. This was consistent with an inability to culture large numbers of S. iniae from the brain of these resistant fish 24 h post-challenge (unpublished data).
The splenic parenchyma is normally comprised of a thin fibrous capsule that surrounds areas of red and white pulp supported by thin smooth muscle trabeculae (Zapata 2024). The red pulp is organized into cords and sinusoids, whereas the white pulp contains ellipsoids characterized by terminal arterioles sheathed by reticular cells, endothelial lined blood-filled channels, and resident macrophages or pigmented macrophage aggregates. Lymphoid cells are at the periphery of the ellipsoids and macrophages (Zapata 2024). In the S24 tilapia, the spleen of all five fish were hypercellular and the periarteriolar sheaths were expanded by moderate to high numbers of histiocytic cells admixed with fewer neutrophils and rare lymphocytes (Figures 2, 3). Within the cytoplasm of the histiocytes and macrophages, there were numerous gram-positive bacterial cocci. Extracellular coccoid bacteria were also observed within vessels and associated with intravascular fibrin thrombi. In one fish, there was segmental full thickness mural necrosis of a large vessel that was surrounded by inflammation, gram-positive bacteria, and cellular debris (Figures 4A and 4B). Alternatively, the spleens of fish from groups S0, R0, and R24 had minimal microscopic changes (Figures 2, 3) with no microscopically detectable bacteria. One fish within the R24 group demonstrated an increase in the erythrocyte population expanding the red pulp (congestion), but a pathological cause was not observed in the section and the distribution of erythrocytes may have been associated with a normal physiological response.
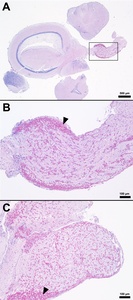
Histopathological changes were observed in the central nervous system of all five S24 fish and were characterized by multifocal regions of mildly to severely inflamed meningeal layers. The pattern of inflammation was usually randomly distributed but was occasionally vasocentric. There was a continuum of changes starting with mild expansion of the meninges by mononuclear cells or hemorrhage in S24 samples (Figures 2 and 3) that transitioned to more severe changes characterized by inflammation, hemorrhage, fibrin, edema, cellular debris, and coccoid organisms (Figure 3). The inflammatory components were predominated by histiocytic cells mixed with fewer lymphocytes and rare eosinophilic granular cells. Hemorrhage and edema were also present in the inflamed areas. Alternatively, the S0, R0, and R24 groups had minimal changes affecting the meninges (Figures 2 and 3). Two R24 fish exhibited rare regions showing meningeal expansion by low numbers of histiocytic cells admixed with fewer lymphocytes. In one R24 fish, there was also a focal submeningeal nodular aggregation of lymphocytes in the optic tectum. No bacterial organisms were observed in the R24 group.
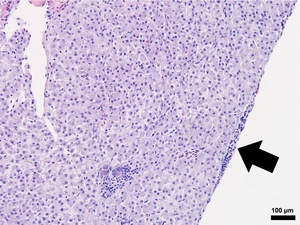
Composed of hematopoietic and lymphoid tissue supported by an interstitium of sinusoids lined by phagocytes, the anterior kidney exhibited normal morphology in the S0, R0, and R24 fish specimens (Figures 2 and 3). Alternatively, the S24 fish had increased numbers of histiocytes that expanded the parenchyma, lymphocytes were reduced in number, and hematopoietic cells were prominent and hyperplastic (Figure 3). The liver samples had variable amounts of hepatocellular intracytoplasmic glycogen throughout the groups. The liver for S24 and R24 tilapia lacked pathological changes except for two fish from each group, that had rare foci of subcapsular and peripancreatic lymphocytes mixed with fewer eosinophilic granular cells (Figure 5).
One unexpected finding among the brain samples of S0, R0, S24, and R24 fish was the observation of variable numbers of eosinophilic granular cells (EGCs) multifocally infiltrating and expanding the meninges of the telencephalon, diencephalon, mesencephalon, and metencephalon (Figure 6). The most significant areas affected by EGC infiltration were located within the optic nerve and adjacent meninges (Figures 6B, 6C). Specific areas infiltrated by the eosinophilic granular cells included the cranial optic tectum, hypothalamus, brainstem, and perivascular cuffs throughout the meninges. Rare peripheral nerves also contained the eosinophilic granular cells in low numbers. This cell type has properties akin to mast cells in higher vertebrate hosts (Romano, Oliveira, and Pedrosa 2021). ECGs have a range of unknown and known functions in teleost fish including production of vasoactive substances, antimicrobial peptides, anti-infectious agents, and inflammatory mediators (Sfacteria, Brines, and Blank 2015). While these cells can be seen in low numbers of vital tissues in clinically healthy fish host (gill, tegument, central nervous system, optic nerve, reproductive tract, and gastrointestinal mucosa), they can also increase with the presence of underlying infection or antigenic stimulation (Camus 2001). A specific cause of the eosinophilic granular cell presence in the present study was not observed but was not associated with S. iniae infection. The ECGs noted in these tilapia are considered an incidental finding and possibly associated with normal anatomic variation for this species.
The host mechanisms responsible for the lack of S. iniae associated lesions and observable gram-positive cocci in the resistant fish are unknown, but of great interest. In our GWAS research, a few potential candidate genes behind the QTL’s were identified that may have roles in innate immunity or host-pathogen interactions such as ccr10 (chemokin receptor 10), rnf213 (RING finger domain), grid1b (glutamate receptor, ionotropic, delta 1b), among others (Vela-Avitúa et al. 2023). The ccr10 gene and glutamate receptor genes like the grid1b gene are upregulated in tilapia following S. iniae challenge (Zhu et al. 2015; Qiang et al. 2017). The rnf213 gene is also upregulated in other fish species following bacterial challenges (Zhang et al. 2022). Our laboratory is currently performing transcriptome studies of the resistant and susceptible tilapia families following experimental S. iniae challenge to close this knowledge gap.
The research conducted through our selective breeding program also suggests that different immune mechanisms are likely involved in resistance to S. iniae, S. agalactiae, and F. orientalis. Our research has demonstrated that there are no genetic correlations between the resistance of tilapia to these different pathogens (Shoemaker et al. 2017; LaFrentz et al. 2020; Shoemaker et al. 2022) as well as no genetic correlations between disease resistance and growth of the fish (LaFrentz et al. 2020; Shoemaker et al. 2022). If similar immune mechanisms were involved in resistance to the different pathogens, we would suspect a positive genetic correlation (i.e., breeding for resistance to one pathogen results in increased resistance to another pathogen), but thus far none have been demonstrated. Therefore, multi-trait selection is used to increase disease resistance to multiple pathogens as well as growth in tilapia.
In summary, evaluation of the organ samples from tilapia selectively bred for resistance or susceptibility to S. iniae revealed clinically significant inflammation in the anterior kidney, spleen, and brain of the susceptible fish 24 h post challenge. Although the number of fish used in this study was small, significant and relevant changes in the vital organs of susceptible fish compared to resistant fish provide evidence that there is a direct link between genetic makeup and resistance to infection. The lesions observed in susceptible fish were consistent with classic S. iniae infection including histiocytic meningitis, splenitis, and nephritis. The marked reduction in disease severity and lack of observable S. iniae in the tissues from resistant tilapia helps to explain the increased survival of these fish following the experimental challenge.
Acknowledgements
This project was supported by a Material Transfer Agreement – Cooperative Research and Development Agreement (MTA-CRADA No. 58-6010-6-005) and USDA-ARS CRIS Project No. 6010-32000-027-00D, Integrated Research to Improve Aquatic Animal Health in Warmwater Aquaculture). The authors acknowledge the excellent technical assistance of Hideyoshi Segovia Uno and Carlos Lopez of Spring Genetics and Megan Justice of USDA-ARS. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture.


__spleen_(c__d)__and_anterior_kidney_(e__f)_from_susceptib.jpeg)
__spleen_(c__d)__and_anterior_kidney_(e__f)_from_resist.jpeg)

__spleen_(c__d)__and_anterior_kidney_(e__f)_from_susceptib.jpeg)
__spleen_(c__d)__and_anterior_kidney_(e__f)_from_resist.jpeg)