INTRODUCTION
The turbot, Scophthalmus maximus, is an economically important flatfish species distributed across the Atlantic, Baltic, Mediterranean, and Black Sea. Overfishing continues to threaten wild turbot populations annually. Over 30 years, global catch volumes have significantly declined due to reduced wild turbot stocks (Aydın et al. 2022). As wild catch alone cannot meet the human consumption need, turbot aquaculture has continued to expand. Over the past 40 years, global turbot aquaculture production has increased substantially, with China, Spain, and Portugal being the primary production centers (FAO 2021).
Turbot is a carnivorous species that inhabits benthic or near-bottom marine environments and holds considerable potential for aquaculture along the Black Sea coast. For nearly three decades, the Central Fisheries Research Institute (SUMAE) has been cultivating this species for experimental and stock enhancement purposes. Under a marine-based rearing system, approximately 10,000 larvae are produced annually. These juvenile turbots are tagged and subsequently released into various locations along the Turkish Black Sea coastline (Aksungur et al. 2007).
Bacterial and viral pathogens cause significant economic losses in turbot aquaculture. In recent years, due to the intensification of aquaculture conditions, disease outbreaks and mortalities caused by pathogens have become a critical issue for the global turbot industry (Feng et al. 2020). Aeromonas salmonicida (Wang et al. 2020), species of Vibrio, Aeromonas, and Pseudomonas (Totoiu et al. 2023), and the Viral Hemorrhagic Septicemia Virus (Albayrak et al. 2018) have been identified as pathogenic bacteria and viruses causing mortality in turbot.
Overall, this study represents the first report focusing on disease issues encountered in turbot aquaculture in Türkiye. It provides valuable insights into bacteria’s ecological distribution and pathogenicity, serving as a foundation for future aquaculture practices and research.
MATERIALS and METHODS
All turbot procedures were carried out per National and Institutional Animal Welfare Guidelines. Ethical approval for the study was granted by the Local Ethical Committee of the SUMAE Türkiye, under protocol number 325.04.02-19. The turbots used in this study were cultured in a flow-through seawater system under SUMAE’s supervision in Trabzon, Türkiye. In 2024, the health status of adult turbots in the facility was monitored monthly. Approximately 10 moribund or recently deceased turbots (weighing 40–50 g) exhibiting clinical signs of disease were aseptically collected each month and transported to the fish diseases laboratory for bacteriological analysis. Water parameters during each sampling event were recorded: temperature 13-22°C, pH 7-7.4, and salinity 18 ppt.
Bacterial examination
Approximately 10 juvenile turbots were subjected to macroscopic examination each month, particularly concerning the fin bases, skin, and ocular areas. Sterile swabs were used to inoculate samples from the head-kidney, liver, and spleen onto TCBS (Thiosulfate Citrate Bile Sucrose Agar; Merck, Germany) and TSA (Tryptic Soy Agar; Merck, Germany) supplemented with 1.5% NaCl. The inoculated plates were incubated aerobically at 25°C for 48 hours. Following incubation, bacterial colonies with smooth morphology were sub-cultured onto TSA to obtain pure isolates. Biochemical characterization of the pure isolates included Gram staining, cytochrome oxidase activity, catalase activity, and motility assays, performed according to the protocols described previously (Ture, Altinok, and Alp 2018).
DNA extraction, PCR assay, and sequencing
The genomic DNA of all bacterial isolates was extracted using the QIAmp DNA Mini Kit (Qiagen, Germany), per the manufacturer’s instructions. The concentration and quality of DNA were measured using a NanoDrop spectrophotometer (ND 8000; Thermo Fisher Scientific, USA), and the DNA concentrations of each isolate were adjusted to 50 ng/μL. The template DNA was stored at -20℃ until use.
The partial 16S rRNA gene of the bacterial isolate was amplified using the forward primer 63f (5-CAG GCC TAA CAC ATG CAA GTC-3) and the reverse primer 1387r (5-GGG CGG WGT GTA CAA GGC-3), following the protocol described by Marchesi et al. (1998). PCR amplification was performed in a thermal cycler (Applied Biosystems, Germany) using the AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific, USA). The resulting PCR products were analyzed by electrophoresis on a 1.5% agarose gel prepared in 1×Tris-Acetate-EDTA (TAE; AppliChem, USA) buffer. A 100-bp DNA ladder (Bioneer, Korea) was used as a size standard to estimate DNA fragment sizes.
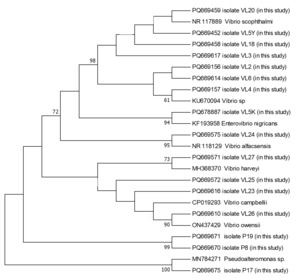
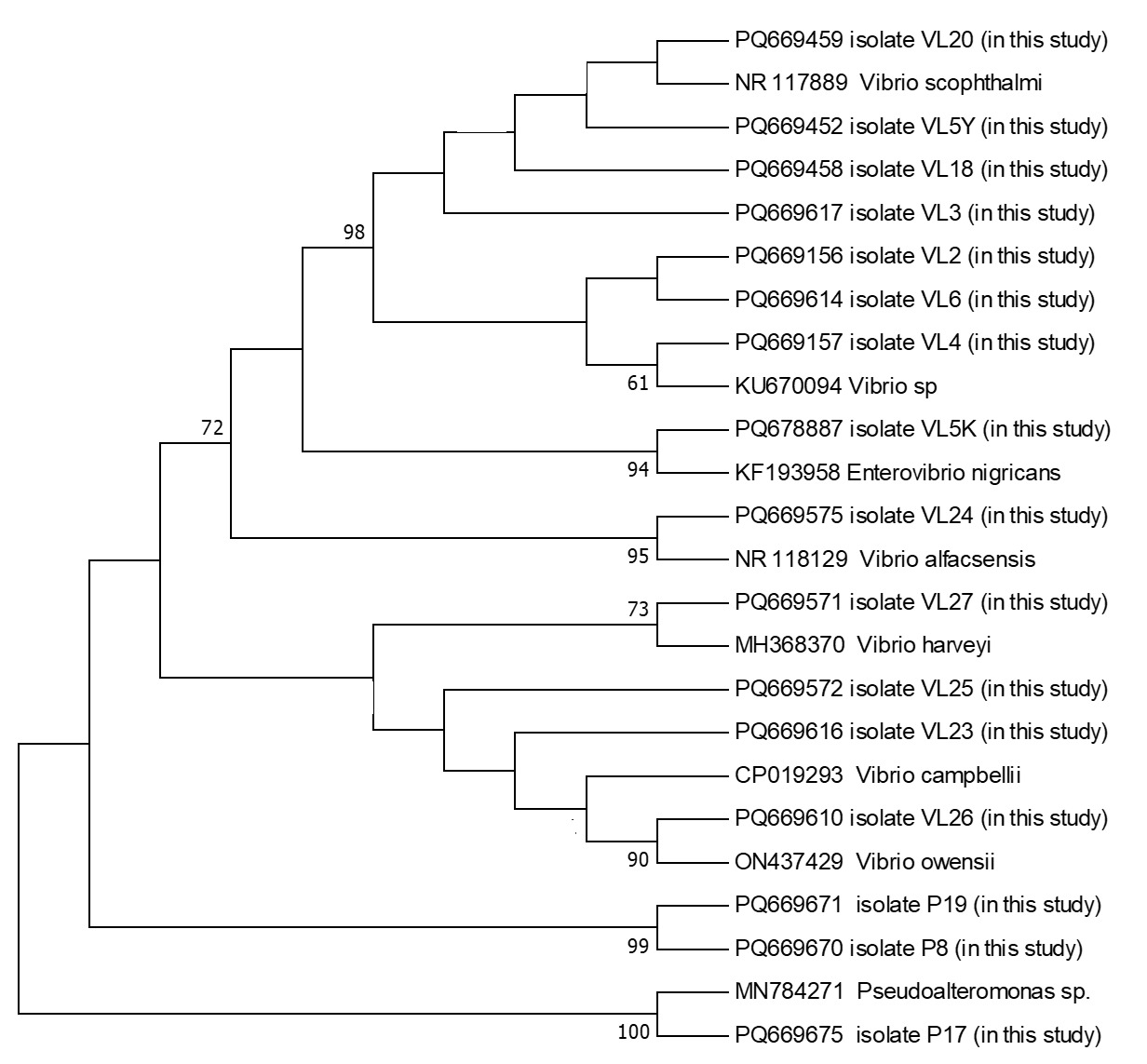
The PCR products were subsequently sequenced on an ABI 3500 Genetic Analyzer (Applied Biosystems, USA) using the BigDye Terminator Cycle Sequencing Kit v3.1. Sequence alignment and editing were performed using BioEdit software (Hall 1999). The sequences were aligned using the ClustalW algorithm (Thompson, Higgins, and Gibson 1994), and low-quality sequences were excluded. The finalized sequences were submitted to GenBank database and compared with publicly available sequences using the BLAST algorithm. A phylogenetic tree based on the 16S rRNA gene sequences was constructed using the maximum likelihood method in MEGA 11 (Tamura et al. 2011), with 1000 bootstrap replicates for statistical validation.
Antimicrobial susceptibility testing
The antimicrobial susceptibility of the isolated bacterial strains was evaluated using the disk diffusion method on Mueller Hinton Agar (MHA, Merck). The testing procedure and result interpretation were performed by the guidelines established by the Clinical and Laboratory Standards Institute (CLSI 2014). The zone diameters of the bacteria were interpreted based on the standard interpretive criteria (breakpoints) established for Enterobacteriaceae. The following antimicrobial discs (Oxoid, UK) were utilized: oxolinic acid (5 μg), penicillin G (10 units), enrofloxacin (5 μg), florfenicol (30 μg), amoxicillin/clavulanic acid (30 μg), erythromycin (15 μg), sulfamethoxazole/trimethoprim (25 μg), and tetracycline (30 μg).
RESULTS
During the 2024 production season, a group of juvenile turbots showed disease symptoms including exophthalmia, ulcers around the dorsal area, petechial hemorrhages at the base of fins, loss of appetite, and abdominal bloating. Isolated bacteria were comma-shaped, gram-negative, and motile. Additionally, cytochrome oxidase and catalase showed a positive reaction. These biochemical properties resembled bacteria of the genus Vibrio, which are frequently isolated from marine environments. The PCR amplification of the 16S rRNA gene from the bacterial strains yielded a product of approximately 1300 base pairs. Sequence analysis demonstrated that the isolates share about 99.00% similarity with the 16S rRNA gene sequences of related bacterial genera or species in the GenBank database. The 16S rRNA gene sequences generated from the bacteria were deposited in GenBank under the accession numbers listed in Table 1. Furthermore, phylogenetic analysis based on the 16S rRNA gene sequences revealed that our isolates clustered closely with members of the Vibrio, Enterovibrio, and Pseudoalteromonas genera (Figure 1).
In total, 16 bacterial isolates representing 8 different species from 3 distinct genera were obtained during this study (Table 2). No notable differences in external clinical signs were observed among fish infected with different bacterial species. Likewise, seasonal variation in the bacterial isolates from turbot was minimal, with Vibrio consistently emerging as the dominant genus.
Antimicrobial susceptibility testing revealed that all isolated bacteria were sensitive to enrofloxacin, florfenicol, and gentamicin. However, varying degrees of resistance were observed among the isolates, with resistance rates of 62.5% for penicillin, 50% for amoxicillin, 43.75% for sulphamethoxazole, 37.5% for tetracycline, 18.75% for erythromycin, and 18.75% for oxolinic acid. The highest resistance rate was recorded for penicillin.
DISCUSSION
From past to present, developed countries have prioritized the production of high-protein quality products for human consumption. Within this scope, turbot, a globally distributed marine fish species of significant economic importance, is widely cultivated due to its high market value. In Türkiye, turbot farming has become an important sector within the aquaculture industry, with production volumes steadily increasing yearly (Zengin, Gumus, and Bostanci 2006). This study focused on monthly examinations of bacterial pathogens in reared juvenile turbot. The investigation identified 16 bacterial isolates representing 8 species and 3 genera.
Vibrio species lead to substantial economic losses, especially among cultured fish species in seawater systems. Skin lesions, hemorrhages, and septicemia typically characterize the disease. Fish and shellfish, regardless of age and across a wide range of species, are highly susceptible to infections caused by Vibrio spp. The infection’s progression depends on the bacteria’s virulence factors, although host defense mechanisms can influence disease outcomes (Manchanayake et al. 2023). Vibrio species are predominantly part of the normal microbiota in temperate marine environments and are also present in the intestinal flora of many aquatic organisms. Among Vibrio species, there are strains with pathogenic, opportunistic pathogenic, and commensal characteristics. Moreover, commensal and opportunistic species may cause mortality in fish depending on environmental conditions (Egerton et al. 2018). Numerous studies have focused on Vibrio species in turbot aquaculture areas. Vibrio splendidus and Vibrio campbellii (Diggles et al. 2000), Vibrio scophthalmi (Cerdà-Cuéllar and Blanch 2004), Vibrio harveyi (Yuan et al. 2021) and Vibrio alfacsensis (Hu et al. 2024) are the predominant Vibrio species isolated. The common feature of all these studies is the reporting that juvenile turbots affected by Vibrio infections exhibit distinct clinical symptoms, including loss of appetite, swimming disorder, abdominal distension caused by yellowish fluid accumulation in internal organs, and hemorrhagic lesions at the fin bases. In our study, juvenile turbots were monitored for bacterial diseases for 1 year. In the study where Vibrio species were determined as the dominant bacteria species, symptoms such as hemorrhage, skin redness, and fluid accumulation in the abdominal cavity were observed in fish, similar to the literature.
In recent years, enrofloxacin, florfenicol, and sulfamethoxazole have been commonly used in the treatment of bacterial fish diseases in Türkiye (Ture, Altinok, and Alp 2018). According to the results of antimicrobial susceptibility testing performed in this study, the isolated bacterial strains were susceptible to enrofloxacin and florfenicol, while exhibiting intermediate resistance to sulfamethoxazole.
A study conducted in the marine areas of Romania assessed the health conditions of turbot fish, the presence of pathogenic agents, and their effects on the fish. Predominantly, pathogens from the genera Vibrio, Aeromonas, and Pseudomonas were identified in turbot. Numerous skin diseases and neoplasms, potentially related to fishing activities, were also detected (Totoiu et al. 2023). No neoplasms or similar skin diseases have been detected in juvenile turbot produced at SUMAE; however, such conditions are occasionally observed in fish older than five years. It should be considered that the formation of neoplasms may not be solely attributed to anthropogenic factors, as bacterial and viral agents could also play a significant role in disease development.
As a result, in the study where bacterial pathogens of juvenile turbot were examined monthly, 16 bacterial strains representing 8 species belonging to 3 different genera were isolated. Bacterial isolates were taxonomically classified into the genera Vibrio, Enterovibrio, and Pseudoalteromonas; these included Vibrio sp., V. scophthalmi, V. alfacsensis, V. harveyi, V. campbellii, V. owensii, Enterovibrio nigricans, and Pseudoalteromonas sp. All of these were previously reported as opportunistic or potentially pathogenic.
Funding Information
The Republic of Türkiye, the Ministry of Agriculture and Forestry, and the General Directorate of Agricultural Research and Policies funded this project (Project no: TAGEM/HSGYAD/B/21/A5/P4/2505).
Conflict of Interest
The authors declare that there are no conflicts of interest.
Author Contribution
Mustafa Türe: Conceptualization, investigation, methodology, writing-original draft, and writing-review. Esen Kulaç Polat: Investigation, project administration, methodology and writing-review. Atife Tuba Beken: Supervision, investigation, project administration, methodology, funding acquisition and writing-review. İlyas Kutlu: Investigation.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethical approval
Ethical approval was mentioned in the text.