Introduction
Cyprinid herpesvirus 3 (CyHV-3), recently renamed Cyvirus cyprinidallo3 (ICTV 2022), and also known as Koi Herpesvirus, is a highly pathogenic virus responsible for the Koi Herpesvirus Disease (KHVD) in common carp and ornamental koi Cyprinus carpio. CyHV-3 belongs to the genus Cyvirus, family Alloherpesviridae and order Herpesvirales (Waltzek et al. 2005). CyHV-3 causes massive mortalities in common carp farming after 5–7 days and has been highlighted as a serious threat to aquaculture and wild populations (Boutier et al. 2015). Since its emergence in 1996, CyHV-3 is present in almost all continents (Rakus et al. 2013). Several factors, such as virus virulence, association with bacterial infections, host condition and water temperature, have been shown to determine the outcome of CyHV-3 infection (Omori and Adams 2011; Padhi et al. 2019; Yi et al. 2014). In most farming settings, preventive efforts against CyHV-3 infection rely on avoiding exposure to the disease by applying efficient biosecurity measures as well as surveillance programmes (Amin et al. 2018).
CyHV-3 is a structurally complex virus possessing the largest genome (295 kb) known among alloherpesviruses, which encodes 156 putative genes, or open reading frames (ORFs) (Aoki et al. 2007). The large size of its genome and the restricted number of full-length sequences obtained from in vivo samples has become an obstacle to understanding its evolution. Using specific genetic markers, Sunarto et al. (2011) hypothesized that Indonesian isolates all originate from a single introduction, followed by a rapid spread over the entire country.
Several ORFs have been investigated for their specific role in the virus biology, especially its pathogenicity (Liu et al. 2020; Gao et al. 2023). This is the case of ORF150, which belongs to the really interesting new genes (RING), potentially involved in virus-host interaction by hijacking the cellular E3 ligases (Heilingloh et al. 2014). CyHV-3 ORF150 may down-regulate carp’s inflammatory response to enhance viral proliferation (Klafack et al. 2022). Of interest, a deletion of 1363 bp in ORF150 was shown to be associated with a loss of virulence during challenge tests and served for the elaboration of a vaccine candidate (Klafack et al. 2022). Genomic comparisons of the same cell culture-propagated isolate revealed a dynamic accumulation of structural variations in ORF150 along successive passages on CCB cells (Fuandila et al. 2022). These comparisons showed that CyHV-3 can evolve rapidly during infectious cycles in cell culture (without selection pressure) and that structural variations are an essential component in the evolutionary process of this virus. These results raised the question of whether such structural variations, particularly the deletion in ORF150, also occur in vivo to modulate the virus pathogenicity. To address this question, 43 common carp farms in Indonesia’s enzootic and non-enzootic areas were investigated. Both qPCR-based and sequencing analyses were used to detect the presence of this deleted variant in the associated CyHV-3 strains and identify other potential structural variations in the ORF150. This study was also an opportunity to evaluate the prevalence of CyHV-3 in Indonesia’s biggest carp production areas.
Materials and Methods
Field sample collection
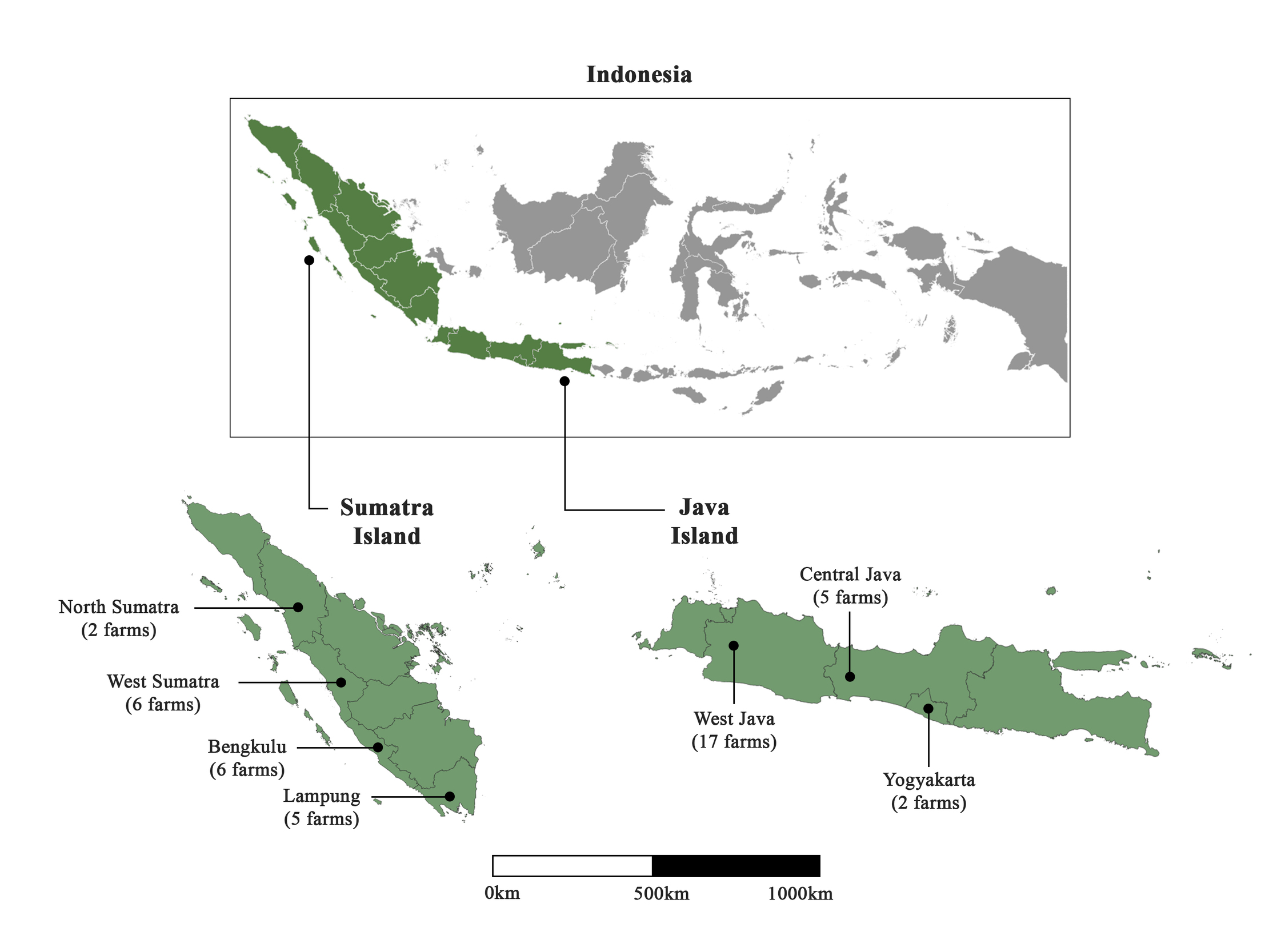
Sample collection of common carp (C. carpio) was carried out between January and May 2022 in 43 farms distributed in 7 provinces throughout Java and Sumatra islands (Figure 1). Fish weighing 10–500 g were harvested from the ponds, and a small piece of gill was taken out, placed in 95% ethanol and kept at -20°C upon arrival to the lab. Fish were then released back into their pond. Information on the collection date, area, size of the fish and number of fish collected at each site are gathered in Supplementary Table 1. In the present study, 37 fish pond farms under various ownerships, and six floating net cage farms, were collected. Sampling farms corresponded to both enzootic and non-enzootic areas of Indonesia (according to fish farmers), and sampling occurred during both epidemic and non-epidemic periods (Table S1). Fish were classified into three groups based on the level of observed clinical signs (Monaghan et al. 2014): 1 (severe clinical signs), 2 (intermediate clinical signs) and 3 (absence of clinical signs). A total of 236 gill samples were collected. Water parameters such as dissolved oxygen, temperature and pH were also measured at a depth of 40 cm. Dissolved oxygen varied between 3.8 and 4.75 mg L-1, and pH between 6.5 and 8.2. Temperature ranged from 19°C to 30.5°C in Java, and from 27°C to 31.5°C in Sumatra.
DNA Extraction
Viral genomic DNA was extracted using the Nucleospin virus extraction kit (Macherey Nagel, Germany), following the manufacturer’s instructions. Briefly, 10-15 mg of gills were grounded using a pestle in a 1.5 mL microtube, and then tissue homogenates were centrifuged at 4000 x g for 10 min at 25°C. Supernatants were transferred into a new 1.5 mL tube to add proteinase K and RNase A solutions. After several wash steps, DNA was eluted with 60 μL of distilled water provided in the kit. Purity of the obtained DNA was verified using a Nanodrop spectrophotometer.
qPCR assays
All 236 samples were tested for i) CyHV-3 carriage and ii) the potential presence of the 1.3-kb deletion in ORF150 (Klafack et al. 2019). For CyHV-3 detection, the improved Sph I-5 primers (Yuasa et al. 2005) were used, and DNA was amplified exactly as described in Avarre et al. (2012). Each sample was run in triplicate, and each run contained several no-template controls. Samples were classified into three categories: + when the three replicates had a cycle of quantification (Cq) lower than 30 and a single melting peak; +/- when only one or two replicates fulfilled the above criteria; - when the three replicates were negative (Cq >30). To evaluate the presence of the deletion in the Indonesian viral strains, two primer sets were used, as described in Klafack et al. (2019): one set targeting the ORF150 of CyHV-3 (nt 259,965 – 260,110 according to GenBank #AP008984.1), and the other located in the putative deletion (nt 258,548 – 258,659 according to GenBank #AP008984.1). Primer sequences are displayed in Table 1. Amplification reactions contained 2.5 μL of 2X SYBR Green I Master mix (Roche), 200 nM of each primer, and 0.5 μl of template DNA in a final volume of 5 μL. qPCR amplifications were carried out in a LightCycler® 480 (Roche). Cycling conditions consisted of an initial denaturation at 95°C for 10 min followed by 45 cycles of amplification at 95°C for 10 sec, annealing at 60°C for 20 sec and elongation at 72°C for 10 sec with a single fluorescence measurement. A melting curve was applied to verify the specificity of the amplified products. It consisted of a denaturation at 95 °C for 5 sec, a renaturation at 65 °C for 60 sec and a heating step from 65–97 °C with a ramp of 0.1 °C per second and a continuous fluorescence acquisition. As tested on cell-culture propagated CyHV-3 strains (not shown), the deletion can be confidently detected under two conditions: the ratio between Cq of the two primer sets is significantly lower than one, and the melting profiles of both amplicons are pure.
Amplicon-based MinIon sequencing
Five positive CyHV-3 samples presenting a high viral load (attested by low Cq values) were subjected to an amplicon-based sequencing. To specifically analyse the whole ORF150 genomic region of CyHV-3 (nt 257,103-260,484 according to GenBank #AP008984), a fragment of ~3.3 kb encompassing the whole ORF150 was amplified with a specifically designed primer pair (Table 1). PCR was performed with GoTaq G2 DNA Polymerase (Promega) in a final volume of 40 μL containing 1 μL of total genomic DNA, 10 μL of Taq buffer, 5 μL of 2-mM dNTPs, 2.5 μL of MgCl2 and 1.25 μL of each primer (10 μM). Cycling conditions were as follows: initial denaturation at 95°C for 10 min, 40 cycles of 95°C for 10 sec, 60°C for 20 sec, 72°C for 3 min, and final extension at 72°C for 5 min. Two rounds of PCR using the same primer pair were necessary to obtain an amount of amplicons sufficient for sequencing. The size of the amplicons was verified by gel electrophoresis with 1% (w/v) agarose (Dutscher Scientific) in Tris-acetate EDTA buffer.
Single amplicon bands of the expected size (3.3 kb) were purified using 1X Agencourt AMPure XP beads. The purity of the obtained DNA was tested using the NanoDrop™ One spectrophotometer (ThermoFisher Scientific). The quantity of DNA was estimated using the Qubit dsDNA High sensitivity kit. DNA libraries were prepared using the Rapid Barcoding kit (SQK-RBK004, Oxford Nanopore Technologies), following the manufacturer’s instructions. For each sample, 400 ng of purified amplicon were adjusted with nuclease-free water to a total volume of 7.5 µL and supplemented with 2.5 µL of Fragmentation Mix RB01-5 (one for each sample). The five barcoded samples were combined in an equimolar ratio and the pooled libraries were sequenced on an R9.4.1 flow cell, using MinKNOW version 0.49.3.7.
Raw fast5 files were recalled using the high-accuracy model of ONT Guppy basecalling software version 6.1.5, and the resulting fastq files were analysed exactly as described in Fuandila et al. (2022). The resulting filtered BAM files served as input data for Sniffles SV caller v4,1 (Sedlazeck et al. 2018). Only SVs ≥ 30 bp supported by at least ten reads were kept in the final VCF files. Sequencing depth was calculated for each sample using the plotCoverage tool implemented in deepTools2.0 tool suite (Ramírez et al. 2016).
Results and Discussion
qPCR analysis using Sph I-5 primers revealed that 165 of 236 sampled fish were positive for CyHV-3. Surprisingly, only 66 fish presented obvious severe clinical signs (Table 2). All of the CyHV-3-negative fish looked healthy. A high number of KHVD-negative fish were found in West Sumatra. Though farms from this region had never experienced CyHV-3 infections, several samples were found positive for the virus. All carp from South Bengkulu, Central Java and Yogyakarta farms looked asymptomatic, yet most tested positive for CyHV-3. Even though the situation was not epidemic during sampling, these areas have experienced CyHV-3 outbreaks in recent years. Some farms from Central Java and Lampung showed both CyHV-3-negative and positive (with a single melting peak) fish. The negative CyHV-3 results can probably be explained by a low level of infection, as already observed (Monaghan et al. 2015; Jin et al. 2020). In contrast, farms from Lampung Bay showed variable levels of CyHV-3 infection, even between very close farms. According to the farmers, this region has frequently experienced CyHV-3 outbreaks. Most of the CyHV-3-positive fish were devoid of clinical signs and had previously experienced KHVD. It is known that common carp exposed to CyHV-3 can become persistently infected with the virus (St-Hilaire et al. 2005).
The vast majority of samples from West Java were associated with severe symptoms and most of them presented a high level of amplification. West Java is known to be enzootic for CyHV-3 and has been reported to experience common carp mortalities up to 100% (Sunarto, Rukyani, et al. 2005; Avarre et al. 2012). Several locations were facing an outbreak onset when sampling occurred. Since abiotic factors are known to influence the outcome of a viral infection (Fabian et al. 2016), water temperature, dissolved oxygen and pH were recorded in most sampled farms. Overall, these water parameters were adequate for carp growth in all systems (Oyugi et al. 2012). Permissive temperatures are generally reported to range from 18 °C to 28 °C (Omori and Adams 2011; Fabian et al. 2015). However, the present study showed that water temperatures in common carp farming frequently exceeded this permissive temperature range, between 28 °C and 31 °C. A previous report highlighted that such high temperatures in Indonesia did not prevent the outbreaks, though they could also contribute to maintaining a high level of asymptomatic carriers (Avarre et al. 2012). Altogether, these results show that CyHV-3 is still highly prevalent across Java and Sumatra islands, regions where the virus spread quickly after its first introduction to Indonesia (Sunarto et al. 2011).
The potential presence of the deletion located in ORF150 (ORF150-del) was investigated in all of the CyHV-3-positive samples. No notable amplification differences were observed between the two primer pairs, suggesting that the ORF150-del haplotype was not present in any sample from Indonesia. To confirm this finding, deep sequencing was realised on the samples that presented the highest viral loads, using the MinIon technology. A sufficient amount of DNA could be obtained for five of them. These samples originated from different farms in West Java: BE-1, BE-2, CRT-1, CRT-2 and DJ-6. The average sequencing depth for the five samples generally ranged from 8,000X to 20,000X. Analysis of the obtained 3.3-kb region revealed a limited number of structural variations (inversions, deletions, duplications, and insertions with a length > 30 nucleotides), all of which supported by a low number of reads (between 11 and 110). This analysis confirmed the absence of the large 1.3-kb deletion in ORF150. In addition, six single nucleotide polymorphisms (SNPs) were recorded in this region with a frequency higher than 5%: five were sample-specific, and one was shared by all samples (Supplementary Table 2). All these SNPs were located within ORF150, and four of them were silent (i.e. had no consequence on ORF150 sequence). The SNP shared by all samples resulted in the change of an arginine for a histidine, with a frequency above 90%. Considering that these two amino acids have the same biochemical properties (they are both basic), it is unlikely that this modification has significant consequences on the protein structure. The last one had a frequency of 23 % and only concerned sample CRT-2. Altogether, these results reveal a low level of variations in this region. This contrasts with what was observed in in vitro samples (Klafack et al. 2019; Fuandila et al. 2022) and suggests that the genomic plasticity of CyHV-3 is much lower in vivo than in vitro (Hammoumi et al. 2016). Genomic variations that are extremely dynamic under in vitro controlled conditions are probably constrained in vivo where environmental conditions and the host immune response represent strong selection pressures. The absence of the deleted form of ORF150 tends to indicate that this SV is not, at least alone, involved in the modulation of CyHV-3 pathogenicity in vivo. However, one cannot exclude that this deletion was not found in asymptomatic fish because of low viral loads, which renders detection challenging. Consequently, further confirmation on a larger set of fish is required. Though still technically difficult because of the high content of host DNA in gill samples, long-read sequencing of the entire viral genome would enable study of SVs along the entire length of the viral genome and identify other regions subject to selection and potentially involved in CyHV-3 virulence.
Acknowledgements
N. N. Fuandila benefited from a CampusFrance fellowship for her PhD thesis, and we are very grateful to the French Embassy of Indonesia. Real-time PCR data were produced on the qPHD platform of Montpellier University, with the support of LabEx CeMEB, an ANR “Investissements d’avenir” program (ANR-10-LABX-04-01). This work benefited from the Montpellier Bioinformatics Biodiversity platform, supported by the LabEx CeMEB. Fish samples were collected following the Indonesian ethical regulations on animal welfare and treated in conformity with the Nagoya protocol.