INTRODUCTION
Lactococcus garvieae is a Gram-positive, coccus-shaped bacterium and the aetiological agent of lactococcosis, a clinically and economically significant disease of fish. The infection is characterized by hyperacute and haemorrhagic septicaemia and has become a serious problem in aquaculture worldwide (Chuah et al. 2016). Typical signs and gross lesions of lactococcosis include lethargy, anorexia, melanosis, exophthalmos, visceral serositis with haemorrhages, severe enteritis, and splenomegaly (Ghittino et al. 2003).
Up until recently, L. garvieae was considered the sole aetiological agent of piscine lactococcosis; however, advanced molecular techniques have revealed that new lactococcal species, L. petauri and L. formosensis, are also implicated in the in the disease etiology. Retrospective analysis have demonstrated that numerous outbreaks attributed to L. garvieae were actually caused by L. petauri or L. formosensis (Egger et al. 2023; de Ruyter et al. 2023; Heckman et al. 2024). Due to their close similarity to L. garvieae in clinical presentation, phenotype and genetics, L. petauri and L. formosensis may be misidentified. Isolates of these three species cannot be differentiated by PCR assays targeting the 16S rRNA gene, and currently, MALDI-TOF does not appear to provide sufficient discriminatory power for reliable identification (Heckman et al. 2024). However, sequencing of the 16S–23S internal transcribed spacer (ITS) region and the DNA gyrase subunit B (gyrB) gene has been recognized as a reliable molecular approach for differentiating Lactococcus species (Ferrario et al. 2013; Stoppani et al. 2023; Barbanti et al. 2024).
Lactococcosis is a major disease in fish, particularly affecting rainbow trout (Oncorhynchus mykiss), the most susceptible species, in which the disease manifests acutely with high mortality rates (Pastorino et al. 2019). L. garvieae can infect a wide range of wild and farmed freshwater and marine fish species, particularly when water temperatures exceed 15°C (Ghittino et al. 2003). It is considered an emerging opportunistic pathogen due to its expanding host range and adaptability to diverse environmental conditions (Akmal, Yoshida, and Nishiki 2022; Francés-Cuesta et al. 2022).
Since the 2000s, aquaculture in Italy has steadily expanded, focusing on European sea bass (Dicentrarchus labrax) and gilthead sea bream (Sparus aurata), the two most farmed species in the Mediterranean basin. The sector has seen a gradual transition from traditional farming systems to coastal and offshore farms (Zoli et al. 2023). In the summer 2023, an outbreak of lactococcosis caused by L. garvieae was reported for the first time in European sea bass (Dicentrarchus labrax) in the Gulf of Follonica (Tuscany, Italy) (Salogni et al. 2024). This Gulf is characterised by a high density of intensive in-shore and off-shore fish farms for gilthead sea bream and European sea bass and represents one of the most important production hubs in Tuscany, accounting for over 50% of national fish production (https://eumofa.eu).
The study describes the first documented outbreaks of L. garvieae in farmed gilthead sea bream (S. aurata).
MATERIALS AND METHODS
Fish sampling and anatomopathological examination
From June 2023 to July 2024, a total of 57 gilthead sea bream (Sparus aurata) were collected from two neighbouring fish farms located in the Gulf of Follonica (coordinates: NW LAT 42°54.470’ – LONG 10°38.000’; NE LAT 42°54.470’ – LONG 10°38.717’; SW LAT 42°53.930’ – LONG 10°38.000’; SE LAT 42°53.930’ – LONG 10°38.717’). The fish were reared both in sea cages (n=53) and in tanks (n=4), with water temperatures ranging from 14,5°C to 25°C. Commercial pellets were used as feed. European sea bass (Dicentrarchus labrax) and greater amberjack (Seriola dumerili) were also present on the farms.
Farm veterinarians reported clinical signs including exophthalmos, erratic lateral swimming, swollen abdomen and haemorrhagic skin lesions accompanied by variable mortality rates.
The fish were submitted for anatomopathological examination following standard procedures at the Istituto Zooprofilattico Sperimentale del Lazio e della Toscana (IZSLT) – Pisa, a public health institute. Samples from spleen, kidney, heart, liver, brain, eye and representative skin lesions were collected for further analysis.
Bacteriological examination
Bacteriological analyses were performed using Sheep Blood Agar (BA;5% sheep blood) plates and Marine Broth (MB) cultures from samples of affected eyes, skin lesions, and internal organs (brain, liver, spleen, kidney, and heart). Culture media were incubated at 25°C and bacterial growth evaluated at 24 and 48 hours. Turbid MB cultures were streaked onto solid media to isolate single colonies. Colonies exhibiting distinct morphologies were subcultured to obtain pure cultures.
Isolates were identified based on phenotypic characteristics including colony morphology, hemolytic activity on BA, Gram staining, motility, oxidase and catalase tests, and microscopic morphology under light microscope. Biochemical profiles of relevant isolates were further characterized by macro-method tests and their halotolerance was assessed using Vibrio Broths (BV) with NaCl concentrations ranging from 0% to 10%.
Isolates presumptively identified as, Lactococcus spp. were tested using the API rapid ID 32 strep system (bioMérieux, France) The manufacturer’s protocol was followed withincubation temperatures adjusted to 25°C and 30°C to ensure accuracy (Vela et al. 2000; Ravelo et al. 2001; Heckman et al. 2024). Resulting identification codes were compared against APIWEB database (bioMérieux, France).
The clindamycin susceptibility test (Elliott and Facklam 1996) was also performed to support phenotypic identification.
Six isolates were further analyzed by MALDI-TOF MS (Microflex LT, Bruker Daltonics; – Library MBT 8468, 2019). Formic acid extraction was applied according to the manufacturer’s instructions to improve identification quality. Identification was considered successful with scores above 2.00.
Total genomic DNA was extracted from the isolates using the QIAamp DNA Mini Kit (Qiagen) according to the the manufacturer’s protocol. DNA quantity and quality were assessed with a NanoDrop spectrophotometer (Thermo Fisher Scientific), and samples were stored at -20°C until further use.
Because the 16S rRNA gene does not provide sufficient resolution to distinguish a L. garvieae from L. petauri (Altinok, Ozturk, and Ture 2022), strain identification was confirmed by two independent end-point PCR assays. The first PCR targeted the 16S-23S rRNA ITS region, using the primers 16S: 5’-GCTGGATCACCTCCTTTCT-3’and 23S: 5’-GGTACTTAGATGTTTCAGTTCC-3’ (Kabadjova et al. 2002), with protocol and cycling conditions following Stoppani et al. (2023). The alignment of the 16S-23S ITS sequences were analyzed for single-nucleotide polymorphism (SNP) patterns enabling discrimination of L. garvieae from closely related species such as L. petauri. . This SNP pattern is consistently found in Italian field strains of L. garvieae (Stoppani et al. 2023) and reference strains of L. garvieae: retrieved from NCBI database LMG 9472 (accession number HM241914), LMG 8162 (accession number HM241916), C1 (accession number MZ146920), 108-33 (accession number MZ146924), 106-30 (accession number MZ146925), 168 (accession number MZ146926), 20,684 (accession number AF225967), and L1-5 (accession number AF225968); and Reference strains retrieved from NCBI database of L. petauri strains: LG4 (accession number CP086401; region: 1911306-1911836), LG26 (accession number CP086595; region 2048225-2048755), B1726 (accession number CP094882; region 1717542-1718072), NHH01_13 (accession number JANHCX010000013; region 2696-3716), LG_SAV_20 (accession number SIVY01000041; region 2699-3719).
The second PCR targeted the the gyrB gene, encoding the DNA gyrase subunit B, which has proven effective for phylogenetic and multilocus sequence typing (MLST) analyses (Shahin et al. 2022). Primers gyrB-F: 5’-CATGCTGGTGGTAAATTTGG-3’, gyrB-R: 5’-GTCATCCATTTCTCCTAAACC-3’ were used, following the protocol described by Barbanti et al. (2024). Amplicons were subjected to in-house Sanger sequencing, chromatograms were manually checked, aligned and consensus sequences generated.
The obtained 16S-23S ITS sequences were compared with data from Stoppani et al. (2023), and gyrB sequences were analyzed via BLASTn searches against the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/).
Antimicrobial susceptibility test
Phenotypic antimicrobial susceptibility of ten L. garvieae strains was determined by disk diffusion and minimum inhibitory concentration (MIC) methods. The Kirby-Bauer (KB) disc diffusion assay was performed against erythromycin (15 μg), oxytetracycline (30 μg), trimethoprim- sulfamethoxazole (1.25 μg/23.75 μg), flumequine (30 μg), florfenicol (30 μg), amoxicillin (25 μg), doxycycline (30 μg), enrofloxacin (5 µg). Testing followed CLSI VET03 and VET04 guidelines. Mueller Hinton agar supplemented with 5% sheep blood was inoculated with the bacterial suspensions standardised to 0.5 McFarland turbidity. Plates were incubated at 22°C for 48 hours.
MICs of the 10 strains were determined using a customized Thermo Scientific micromethod plate. Concentration ranges of antibiotic tested: erythromycin (0.063-32 µg/mL), amoxicillin (0.016-32 µg/mL), florfenicol (0.031-64 µg/mL), flumequine (0.016-32 µg/mL), oxytetracycline (0.008-16 µg/mL), sulfamethoxazole trimethoprim (0.004/0.074-8/152 µg/mL), doxycycline (0.008-16 µg/mL), enrofloxacin (0.004-8 µg/mL). Tests were performed in Cation-Adjusted Mueller Hinton Broth supplemented with 5% lysed horse blood (CAMHB+LHB) and the plates incubated at 22°C for 48 hours, in accordance with CLSI standards.
RESULTS AND DISCUSSION
Of the 57 gilthead sea bream (Sparus aurata) collected between June 2023 and July 2024, 41 (71.93%) were found to be infected with Lactococcus garvieae, with the first isolation of the pathogen recorded in November 2023. The infected fish measured on average 27cm in length (range 17 - 33 cm) and had a mean weight of 450g (range 90g - 560g). The lactococcosis outbreak was characterised by the rapid onset of general clinical signs, including anorexia, disorientation, and erratic swimming. Typical external signs like exophthalmos also known as “pop-eye” (uni- or bilateral), enucleation of the ocular globe, and diffuse reddening or haemorrhages of skin and in the ocular regions were consistent with observations reported by Salogni et al. (2024) in European sea bass (Dicentrarchus labrax) and in previous studies (Vendrell et al. 2006; Meyburgh, Bragg, and Boucher 2017). Furthermore, elevated water temperatures averaging 18°C, reaching a maximum of 25°C, alongside intensive farming conditions, likely contributed as risk factors by increasing fish stress and exacerbating the severity of L. garvieae infections (Soltani et al. 2008; Salogni et al. 2024).
Anatomopathological and histological examinations
Anatomopathological examination of infected fish revealed lesions commonly associated with lactoccosis. Ocular alterations included exophthalmos, hyperaemia, corneal opacity, ulcerative keratitis, orbital collapse, and, in advanced cases, orbital collapse or loss of the eye globe (Fig. 1a-c) in line with previous reports (Vendrell et al. 2006). Other external gross lesions included haemorrhages and diffuse reddening at the base of fins, opercula, and skin (especially in ventral region), as well as skin erosions and pale gills (Fig. 1d-e).
Internally, affected fish displayed severe pericarditis with grey discoloration and cardiomegaly, hepatomegaly with diffuse petechiae, haemorrhages, and yellow to grey discoloration, splenomegaly with miliary whitish nodules, and bowel dilation (Fig. 2). These findings are consistent with previous literature identifying spleen, liver, brain, gut, kidneys, and heart as primary target organs affected in L. garvieae infections (Vendrell et al. 2006; Meyburgh, Bragg, and Boucher 2017). Similarly, lesions such as splenomegaly, liver haemorrhage, pericarditis and endocarditis have been reported in other marine species affected by lactococcosis, including European sea bass (Dicentrarchus labrax) and yellowtail (Seriola quinqueradiata), (Kusuda and Salati 1993; Salogni et al. 2024).
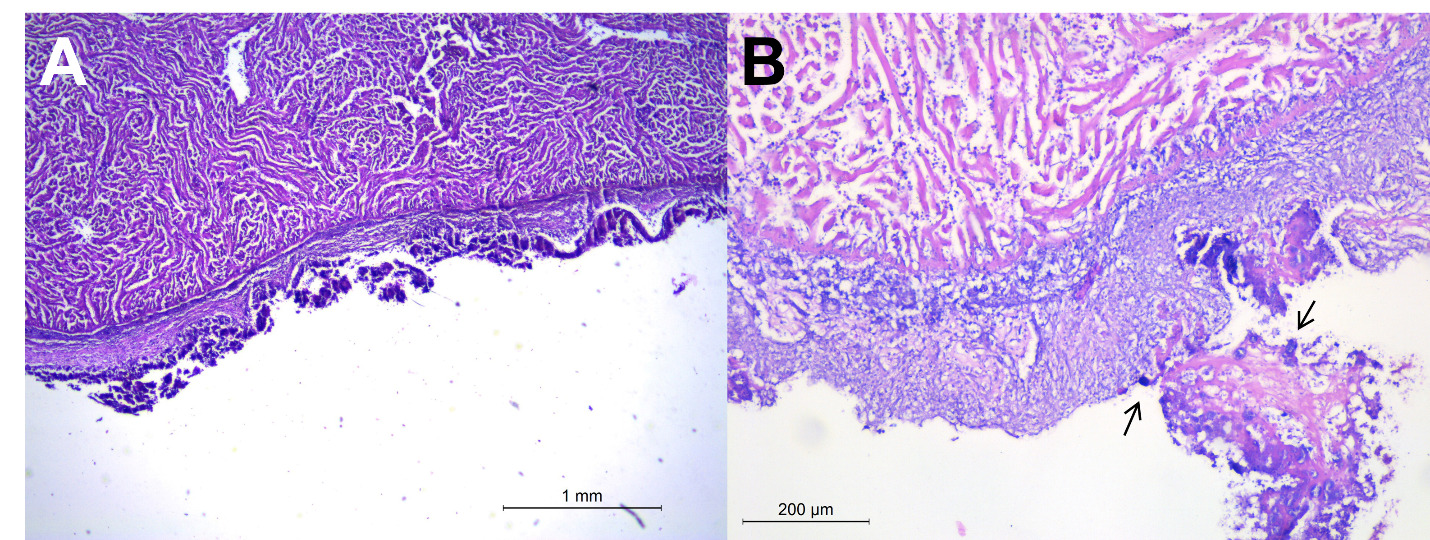
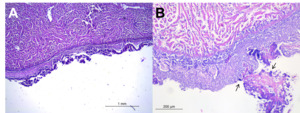
Histological analysis of heart, spleen and liver samples revealed severe fibrinous-necrotic pericarditis and epicarditis, characterised by thickened epicardium with abundant fibrin, histiocytic infiltrate, degenerated inflammatory cells, necrotic debris, and intralesional multifocal bacterial aggregates (Fig.3). Bacterial aggregates were observed both intermixed within the fibrino-necrotic material and, to lesser extent, intracellularly cytoplasm of histiocytes.
The liver, though grossly lacking nodular lesions, presented multiple necrotic foci and large amount of multifocal to coalescing granulomas with necrotic cores (Fig. 4a-b). Similar granulomatous lesions were observed in the spleen, with central core of colliquative necrosis (Fig. 4c-d). Histopathological findings like fibrinous pericarditis, epicarditis, necrotic areas in liver and spleen, are consistent with previous descriptions in lactococcosis-affected fish (Altun et al. 2005; Avci et al. 2014; Salogni et al. 2024). Notably, this study is the first to report granulomatous lesions in the spleen and liver, the latter only histologically, of gilthead sea bream associated with L. garvieae.
Although specific data on feed formulation were not available in this study, the histological evidence of hepatocellular vacuolization in affected fish suggests possible metabolic liver alterations related to dietary imbalance. Similar lesions have been associated with excessive dietary carbohydrate levels in carnivorous marine fish. Gilthead sea bream (Sparus aurata) can utilise carbohydrates up to 20%, but such levels may induce hepatic glycogen accumulation and metabolic stress. The pale livers (Fig. 2b) and observed histological changes (Fig. 4a-b) are consistent with these findings and may indicate liver dysfunction, potentially impairing immune competence and predisposing fish to opportunistic infections such as Lactococcus garvieae (Enes et al. 2006; Moreira et al. 2012; Peres, Santos, and Oliva-Teles 2014).
Identification of L. garvieae: biochemical and molecular methods
Bacteriological analysis of 41 gilthead sea bream revealed pure cultures of L. garvieae from all sampled organs. Colonies on blood agar (BA) were α-hemolytic, small, and grey, tending to become whitish after 48 hours. Isolates were Gram-positive cocci, occurring singly, in pairs or short chains; facultatively anaerobic and non-motile. Salinity tolerance tests, using BV, showed growth at NaCl concentrations of 0-4% for all tested strains, with weak growth observed at 6% NaCl, somewhat differing from Buller’s report of growth at 6 NaCl and weak growth at 6.5% (Buller 2014). These in vitro findings may reflect L. garvieae’s ecological adaptation to marine environments.
The bacterial identification via API rapid ID32 STREP galleries, analyzed using the APIWEB software, yielded codes corresponding to L. garvieae with their respective percentage of identification: 3033 3301 030 (99.5%); 3032 3301 010 (99.5%); 3023 3201 030 (99.8%); 3032 3201 030 (99.8%); 3022 3001 030 (99.9%); 3002 3000 030 (99.9%). No differences in results were observed at the two incubation temperatures; however, some biochemical reactions exhibited more distinct colour development at 30°C making them easier to interpret on the strip. Biochemical tests in the API rapid ID32, including hippurate hydrolysis and acid production from sucrose and tagatose, differentiated L. garvieae from closely related L. petauri being negative for the first and positive for the second (Vela et al. 2024). All strains showed resistance to clindamycin, supporting the differentiation from L. lactis (Elliott and Facklam 1996; Buller 2014).
All the isolates analyzed by MALDI-TOF MS were identified as L. garvieae with scores ranging from 2.01 to 2.36.
The identification of isolates was corroborated using molecular biology approach focusing on the 16S-23S ITS region and the gyrB gene. Molecular characterization based on 16S-23S ITS sequences clustered the isolates with L. garvieae, showing a characteristic SNP pattern as observed in all Italian field and Reference strains (Stoppani et al. 2023): five SNPs and two indel variations that distinguishing between L. garvieae and L. petauri adenine (A) insertion at position g.218_219 insA, 5 SNPs at position g.358G>T, g.440T>G, g.442T>A, g.469C>A, g.478G>T, and thymine (T) deletion at position g.443 (Table 1). BLASTn analysis of gyrB gene sequences performed on Genbank, revealed 100% identity with L. garvieae strain MS210922A (accession number: AP026069).
Antimicrobial susceptibility test (AST)
Disk diffusion (Kirby-Bauer) results (Table 2) showed that L. garvieae isolates exhibited no zones of inhibition for trimethoprim-sulfamethoxazole (1.25 μg/23.75 μg) and flumequine (30 μg), indicating resistance to these antimicrobials. Note that, could not perform a correct interpretation of obtained AST data for lack of the official internationally harmonized AST interpretive criteria also namely Epidemiological cut-off value and Clinical breakpoint; that are currently not available for L. garvieae isolated from aquatic animals.
MIC determinations (Table 3) revealed limited variation among strains, with elevated MICs for trimethoprim-sulfamethoxazole and flumequine. It is clarified that, some MIC breakpoints are available for Lactococcus spp. isolated from humans but the use of these for interpretation may not accurately reflect the in vivo situation for fish.
Erythromycin and oxytetracycline showed the most promising antimicrobial activity based on both MIC and disk diffusion assays.
Therapy
According to the AST results, infected gilthead sea bream in tanks or cages were treated with erythromycin at 75 mg/kg for 10 days via medicated feed. Erythromycin is commonly used for treating lactococcosis in rainbow trout (Agnetti et al. 2008; Serdoz et al. 2011). Initially, mortality and clinical signs decreased rapidly; however, subsequent treatments showed reduced effectiveness, likely due to challenging field conditions. New outbreaks occurred after 3-4 weeks, suggesting limited long-term benefit, potentially linked to the early anorexia onset in infected fish as previously reported (Salogni et al. 2024).
CONCLUSIONS
The isolation of L. garvieae from gilthead sea bream, accompanied by correlated pathological lesions, has not been previously reported in literature. Among the 57 specimens examined between November 2023 and July 2024, 41 showed pure bacterial growth of Gram-positive cocci associated with typical lactococcosis lesions, including ocular and hemorrhagic lesions, pericarditis and endocarditis, splenomegaly and congestion of internal organs. These outbreaks were recorded in the same area where Salogni et al. (2024) described the first case of lactococcosis in European sea bass during summer 2023. At that time, gilthead sea bream within the same farm did not exhibit clinical signs of lactococcosis or increased mortality. Clinical signs, gross pathological findings, and histological examinations indicate that the disease progression is comparable in both marine species.
The isolation of the bacterium in pure culture from all examined organs is consistent with lactococcosis, which typically results in bacteremia and systemic hyperacute infection. While biochemical and proteomic methods provide useful diagnostic support, molecular identification and sequencing of 16S-23S rRNA ITS region and gyrB gene are essential for accurate discrimination of the etiological agent within the Lactococcus genus.
Oral treatment with erythromycin demonstrated limited and variable efficacy. Considering that macrolides are classified by the WHO as critically important antibiotics for human health, their use in aquaculture should be restricted. Therefore, antibiotic therapy appears to be an inadequate long-term control strategy for lactococcosis. Rising mortality rates and difficulties in treating the infection with effective antimicrobials represent a significant concern for gilthead sea bream farming, leading to substantial economic losses and animal health and welfare issues.
Consequently, vaccination and stringent biosecurity measures represent the most effective strategies to control lactococcosis outbreaks. An autogenous inactivated vaccine targeting gilthead sea bream and European sea bass, developed by the pharmaceutical laboratory of IZSLT - Siena, is currently available for Italian fish farmers. However, the feasibility and impact of vaccination programmes will only become appreciable in the medium term.

_severe_exophthalmos__(b)_cornea.tiff)
_haemorrhagic_lesion_of_liver_(w.tiff)

_severe_exophthalmos__(b)_cornea.tiff)
_haemorrhagic_lesion_of_liver_(w.tiff)